Methods
Candidate sgRNAs were scaned genome-wide for N. oceanica IMET1 (NoIMET1v2). Potential off-target sites were searched using cas-offinder with at most 3 mismatches. All sgRNAs which can uniquely target one specific genomic site and without potential off-target sites within 3 mismatches were retained.
Note: To check these sites, please use our genome browser (Link).
Results
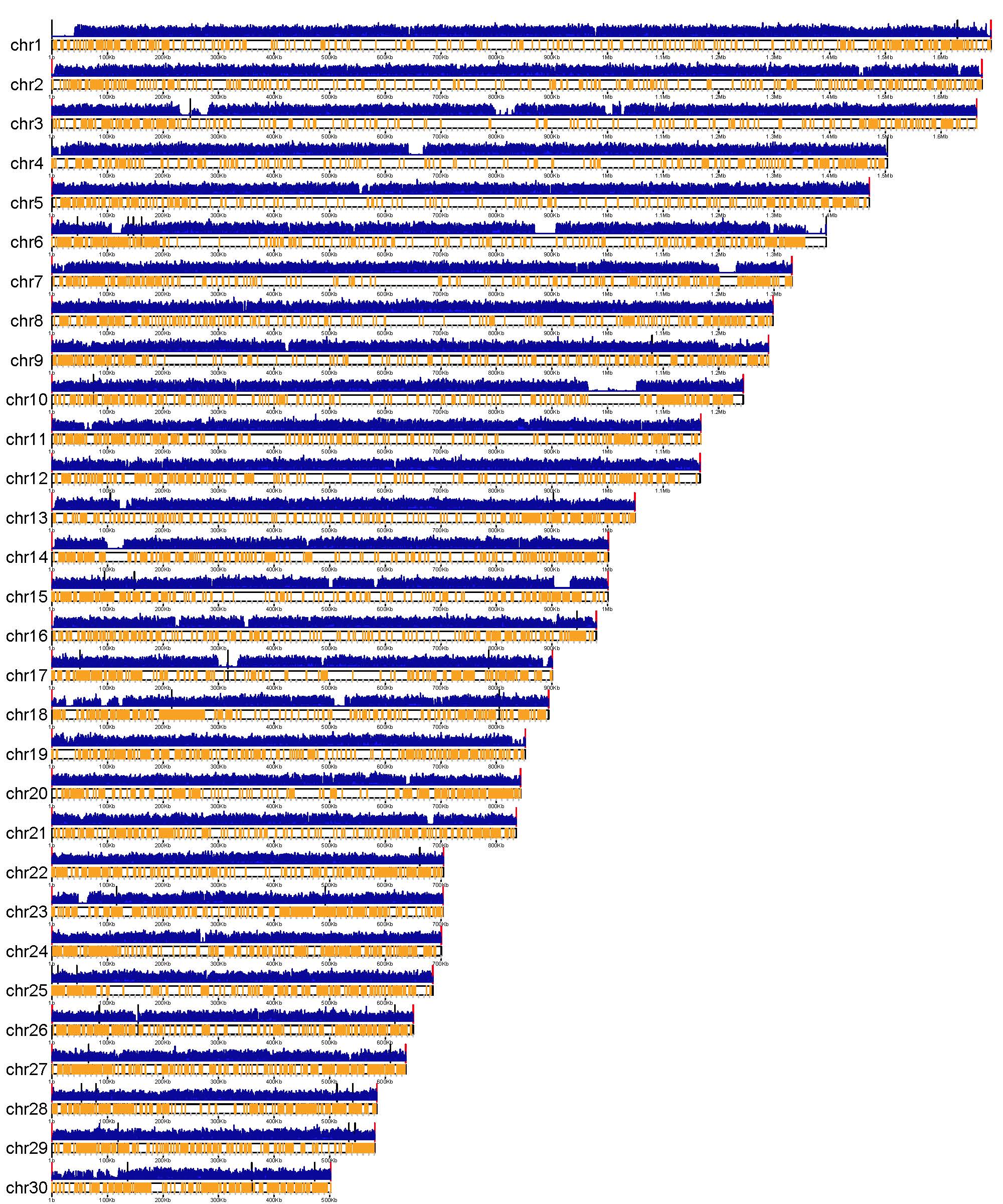
A. abundant and uniformly distributed target-sites can be obtained along the whole genome.
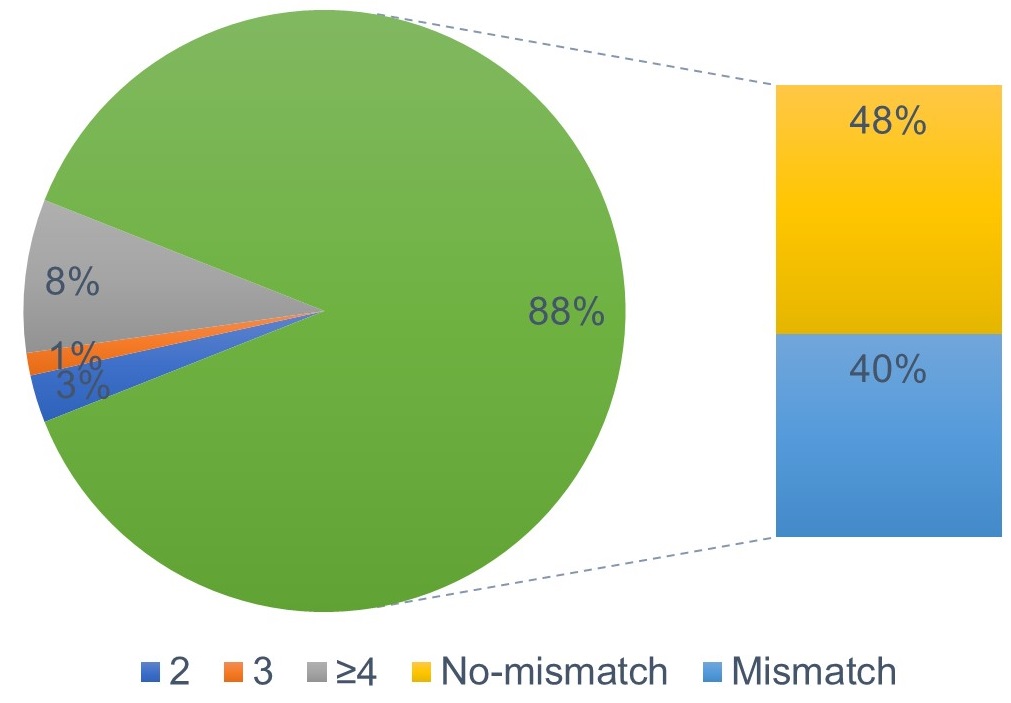
B. 48% sites were uniquely targeted and without potential off-target sites within 3 mismatches.
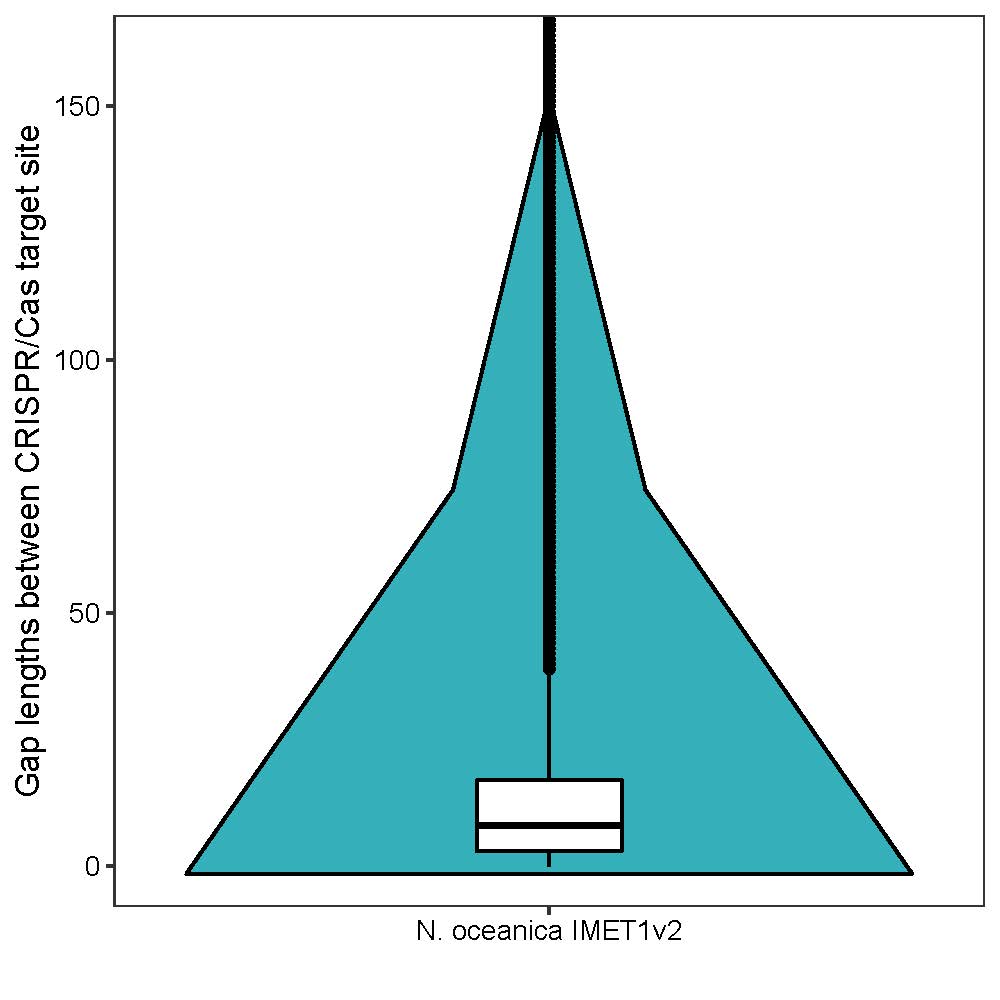
C. Average gap length of adjacent targeted sites was ~ 14 bp.